June 2025
+``` + +**Recommendation:** Fix section ID to match content (`June2025`). + +### People Page - Potentially Outdated + +**File:** `people.html` (950 lines) + +- Team roster may not reflect current active contributors +- Bio information may be outdated + +**Recommendation:** Audit team list for accuracy. + +### Index Page - Google+ Widget (DEAD SERVICE) + +**File:** `index.html:224-227` + +```html + +`, `` | `{"type": "text", "subtype": "heading1"}` |
+| `` | `{"type": "text", "subtype": "heading2"}` |
+| `
` | `{"type": "text", "subtype": "heading2"}` | +| `
Chapter 1: A brief introduction to C. elegans
@@ -166,7 +161,7 @@Educational kits
- + diff --git a/events.html b/events.html index a3c618e..8571755 100644 --- a/events.html +++ b/events.html @@ -16,10 +16,6 @@ - - @@ -206,9 +202,6 @@Computational Neuroscience Meeting 2013
@@ -222,9 +215,6 @@OpenWorm Paris Meet-up
Guest Lecture: Introduction to OpenWorm
Speaker: Matteo Cantarelli. Host: Daniele Giusto - Faculty of Electrical Engineering, University Of Cagliari, Italy
--
-
- Website -
Open Source Brain Kick-Off meeting
@@ -270,9 +257,6 @@OpenWorm Cambridge Meet-up
@@ -383,7 +367,7 @@NeuroML Development Workshop
- + diff --git a/footer-content.html b/footer-content.html index 7eb8de8..e2c1d27 100644 --- a/footer-content.html +++ b/footer-content.html @@ -8,7 +8,7 @@ ·- Code licensed under MIT, documentation - under CC BY + Code licensed under MIT, documentation + under CC BY 3.0.
diff --git a/get_involved.html b/get_involved.html index eda26fc..f6e4439 100644 --- a/get_involved.html +++ b/get_involved.html @@ -14,11 +14,6 @@ - - - @@ -230,7 +225,7 @@
Curious citizen
- + diff --git a/getting_started.html b/getting_started.html index e14e886..b1479c9 100644 --- a/getting_started.html +++ b/getting_started.html @@ -15,11 +15,6 @@ - - - diff --git a/header-content.html b/header-content.html index b3590e1..86331b6 100644 --- a/header-content.html +++ b/header-content.html @@ -39,7 +39,7 @@Open source visualization and simulation platform.
Tweet - -Latest news
Loading tweets...
+Latest news
- + diff --git a/js/main.js b/js/main.js index 827842c..cd6b13b 100644 --- a/js/main.js +++ b/js/main.js @@ -38,9 +38,9 @@ $(document).on('pjax:popstate', function() { loadDonationControls(); } else if (loc === '/index.html' || loc === '/' || loc === '') { - // Twitter widget + // Twitter/X feed widget $('#fb-root').html(''); - $('#tweeter').html('Tweets by @OpenWorm'); + $('#tweeter').html('Loading tweets...
' + title + '
'; + html += '' + dateStr + '
'; + html += '
-
+
diff --git a/migrate_news_to_tumblr.py b/migrate_news_to_tumblr.py
new file mode 100644
index 0000000..ba4d78f
--- /dev/null
+++ b/migrate_news_to_tumblr.py
@@ -0,0 +1,560 @@
+#!/usr/bin/env python3
+"""
+Migrate news.html content to Tumblr with backdating.
+Uses NPF (Neue Post Format) for proper image support.
+"""
+
+import os
+import re
+import json
+from datetime import datetime
+from pathlib import Path
+from bs4 import BeautifulSoup, NavigableString
+from dotenv import load_dotenv
+from requests_oauthlib import OAuth1Session
+
+# Load environment
+ENV_FILE = Path(__file__).parent / ".env.tumblr"
+load_dotenv(ENV_FILE)
+
+BLOG_NAME = "openworm"
+API_BASE = "https://api.tumblr.com/v2"
+SITE_BASE = "https://openworm.org"
+
+# Map month names to dates for backdating
+MONTH_DATES = {
+ "June2025": "2025-06-15T12:00:00Z",
+ "Dec2024": "2024-12-15T12:00:00Z",
+ "May2024": "2024-05-15T12:00:00Z",
+ "June2023": "2023-06-15T12:00:00Z",
+ "September2022": "2022-09-15T12:00:00Z",
+}
+
+MONTH_TITLES = {
+ "June2025": "June 2025",
+ "Dec2024": "December 2024",
+ "May2024": "May 2024",
+ "June2023": "June 2023",
+ "September2022": "September 2022",
+}
+
+
+def get_oauth_session():
+ """Get authenticated OAuth session."""
+ return OAuth1Session(
+ os.getenv("TUMBLR_CONSUMER_KEY"),
+ client_secret=os.getenv("TUMBLR_CONSUMER_SECRET"),
+ resource_owner_key=os.getenv("TUMBLR_ACCESS_TOKEN"),
+ resource_owner_secret=os.getenv("TUMBLR_ACCESS_TOKEN_SECRET")
+ )
+
+
+def fix_url(url):
+ """Convert relative URLs to absolute URLs."""
+ if not url:
+ return url
+ if url.startswith('//'):
+ return 'https:' + url
+ if url.startswith('/'):
+ return SITE_BASE + url
+ if url.startswith('img/') or url.startswith('assets/'):
+ return SITE_BASE + '/' + url
+ return url
+
+
+def get_image_type(url):
+ """Guess image MIME type from URL."""
+ url_lower = url.lower()
+ if '.png' in url_lower:
+ return 'image/png'
+ elif '.gif' in url_lower:
+ return 'image/gif'
+ elif '.webp' in url_lower:
+ return 'image/webp'
+ return 'image/jpeg'
+
+
+def html_to_npf_blocks(html_content):
+ """
+ Convert HTML content to NPF (Neue Post Format) blocks.
+ Returns list of content blocks for Tumblr API.
+ """
+ soup = BeautifulSoup(html_content, 'html.parser')
+ blocks = []
+
+ def normalize_whitespace(text):
+ """Normalize whitespace - collapse multiple spaces/newlines into single space."""
+ return re.sub(r'\s+', ' ', text)
+
+ def extract_text_with_formatting(element):
+ """Extract text and formatting info from an element."""
+ text = ""
+ formatting = []
+
+ for child in element.children:
+ if isinstance(child, NavigableString):
+ # Normalize internal whitespace but check for boundary spaces
+ child_str = str(child)
+ has_leading_space = bool(re.match(r'^\s', child_str))
+ has_trailing_space = bool(re.search(r'\s$', child_str))
+
+ normalized = normalize_whitespace(child_str).strip()
+ if normalized:
+ # Add leading space if original had it and we need separation
+ if has_leading_space and text and not text.endswith(' '):
+ text += ' '
+ text += normalized
+ # Add trailing space if original had it
+ if has_trailing_space:
+ text += ' '
+ elif child.name == 'a':
+ start = len(text)
+ link_text = normalize_whitespace(child.get_text()).strip()
+ text += link_text
+ end = len(text)
+ href = fix_url(child.get('href', ''))
+ if href:
+ formatting.append({
+ "start": start,
+ "end": end,
+ "type": "link",
+ "url": href
+ })
+ elif child.name in ['b', 'strong']:
+ start = len(text)
+ bold_text = normalize_whitespace(child.get_text()).strip()
+ text += bold_text
+ end = len(text)
+ formatting.append({"start": start, "end": end, "type": "bold"})
+ elif child.name in ['i', 'em']:
+ start = len(text)
+ italic_text = normalize_whitespace(child.get_text()).strip()
+ text += italic_text
+ end = len(text)
+ formatting.append({"start": start, "end": end, "type": "italic"})
+ elif child.name == 'br':
+ text += "\n"
+ else:
+ # Recursively handle nested elements
+ nested_text, nested_fmt = extract_text_with_formatting(child)
+ offset = len(text)
+ text += nested_text
+ for fmt in nested_fmt:
+ formatting.append({
+ **fmt,
+ "start": fmt["start"] + offset,
+ "end": fmt["end"] + offset
+ })
+
+ return text.strip(), formatting
+
+ def process_element(elem):
+ """Process a single HTML element into NPF blocks."""
+ if isinstance(elem, NavigableString):
+ text = normalize_whitespace(str(elem))
+ if text:
+ return [{"type": "text", "text": text}]
+ return []
+
+ if elem.name == 'h1':
+ text = normalize_whitespace(elem.get_text())
+ if text:
+ return [{"type": "text", "subtype": "heading1", "text": text}]
+
+ elif elem.name == 'h2':
+ text = normalize_whitespace(elem.get_text())
+ if text:
+ return [{"type": "text", "subtype": "heading1", "text": text}]
+
+ elif elem.name == 'h3':
+ text = normalize_whitespace(elem.get_text())
+ if text:
+ return [{"type": "text", "subtype": "heading2", "text": text}]
+
+ elif elem.name == 'p':
+ # Check if this is primarily an image container
+ img = elem.find('img')
+ if img:
+ src = fix_url(img.get('src', ''))
+ if src:
+ alt = img.get('alt', '')
+ block = {
+ "type": "image",
+ "media": [{
+ "url": src,
+ "type": get_image_type(src)
+ }]
+ }
+ if alt:
+ block["alt_text"] = alt
+
+ # Also check for any text/link accompanying the image
+ result = [block]
+
+ # Check if there's a link wrapping the image
+ parent_link = img.find_parent('a')
+ if parent_link:
+ href = fix_url(parent_link.get('href', ''))
+ if href and href != src:
+ # Add a link block after the image
+ result.append({
+ "type": "text",
+ "text": f"View: {href}",
+ "formatting": [{
+ "start": 6,
+ "end": 6 + len(href),
+ "type": "link",
+ "url": href
+ }]
+ })
+ return result
+
+ # Regular paragraph with text
+ text, formatting = extract_text_with_formatting(elem)
+ if text:
+ block = {"type": "text", "text": text}
+ if formatting:
+ block["formatting"] = formatting
+ return [block]
+
+ elif elem.name == 'img':
+ src = fix_url(elem.get('src', ''))
+ if src:
+ block = {
+ "type": "image",
+ "media": [{
+ "url": src,
+ "type": get_image_type(src)
+ }]
+ }
+ alt = elem.get('alt', '')
+ if alt:
+ block["alt_text"] = alt
+ return [block]
+
+ elif elem.name == 'a':
+ # Standalone link
+ href = fix_url(elem.get('href', ''))
+ text = normalize_whitespace(elem.get_text())
+ if href and text:
+ return [{
+ "type": "text",
+ "text": text,
+ "formatting": [{
+ "start": 0,
+ "end": len(text),
+ "type": "link",
+ "url": href
+ }]
+ }]
+
+ elif elem.name == 'blockquote':
+ text = normalize_whitespace(elem.get_text())
+ if text:
+ return [{"type": "text", "subtype": "indented", "text": text}]
+
+ elif elem.name in ['ul', 'ol']:
+ items = []
+ for li in elem.find_all('li', recursive=False):
+ text, formatting = extract_text_with_formatting(li)
+ if text:
+ block = {
+ "type": "text",
+ "subtype": "unordered-list-item" if elem.name == 'ul' else "ordered-list-item",
+ "text": text
+ }
+ if formatting:
+ block["formatting"] = formatting
+ items.append(block)
+ return items
+
+ elif elem.name == 'kbd':
+ text = normalize_whitespace(elem.get_text())
+ if text:
+ return [{"type": "text", "text": f"`{text}`"}]
+
+ elif elem.name == 'figure':
+ img = elem.find('img')
+ if img:
+ return process_element(img)
+ return []
+
+ elif elem.name == 'div':
+ # Process children of divs
+ result = []
+ for child in elem.children:
+ if not isinstance(child, NavigableString) or str(child).strip():
+ result.extend(process_element(child))
+ return result
+
+ return []
+
+ # Process all direct children
+ for elem in soup.children:
+ blocks.extend(process_element(elem))
+
+ # Clean up: remove empty blocks (or blocks with just whitespace)
+ blocks = [b for b in blocks if (b.get('text', '').strip()) or b.get('type') == 'image']
+
+ # Dedupe images (same URL can appear multiple times due to HTML structure)
+ seen_urls = set()
+ deduped = []
+ for block in blocks:
+ if block['type'] == 'image':
+ url = block['media'][0]['url']
+ if url not in seen_urls:
+ seen_urls.add(url)
+ deduped.append(block)
+ else:
+ deduped.append(block)
+
+ return deduped
+
+
+def parse_news_html(html_path):
+ """
+ Parse news.html and extract individual news items.
+ Returns list of dicts with: section_id, date, title, items
+ """
+ with open(html_path, 'r') as f:
+ soup = BeautifulSoup(f.read(), 'html.parser')
+
+ sections = []
+
+ for section in soup.find_all('section'):
+ section_id = section.get('id')
+ if not section_id or section_id not in MONTH_DATES:
+ continue
+
+ page_header = section.find('div', class_='page-header')
+ if not page_header:
+ continue
+
+ month_title = MONTH_TITLES.get(section_id, section_id)
+ backdate = MONTH_DATES.get(section_id)
+
+ # Find all h3 items (individual news items)
+ items = []
+ current_item = None
+
+ # Get all relevant elements
+ for elem in page_header.find_all(['h1', 'h3', 'p', 'ul', 'ol', 'img', 'blockquote']):
+ if elem.name == 'h1':
+ continue # Skip the main section heading
+
+ if elem.name == 'h3':
+ # Start a new item
+ if current_item:
+ items.append(current_item)
+
+ # Clean up the h3 title (remove numbering like "1) ")
+ title_text = elem.get_text().strip()
+ title_text = re.sub(r'^\d+\)\s*', '', title_text)
+
+ current_item = {
+ 'title': title_text,
+ 'html_parts': [],
+ }
+ elif current_item:
+ # Add content to current item
+ current_item['html_parts'].append(str(elem))
+
+ # Don't forget the last item
+ if current_item:
+ items.append(current_item)
+
+ sections.append({
+ 'section_id': section_id,
+ 'month_title': month_title,
+ 'backdate': backdate,
+ 'items': items,
+ })
+
+ return sections
+
+
+def create_npf_post(oauth, title, npf_blocks, backdate, state="draft", tags=None):
+ """Create a Tumblr post using NPF format."""
+ # Add title as first heading block
+ content = [
+ {"type": "text", "subtype": "heading1", "text": title}
+ ] + npf_blocks
+
+ payload = {
+ "content": content,
+ "state": state,
+ }
+
+ if tags:
+ payload["tags"] = ",".join(tags)
+
+ # NPF supports date parameter too
+ if backdate:
+ payload["date"] = backdate
+
+ response = oauth.post(
+ f"{API_BASE}/blog/{BLOG_NAME}.tumblr.com/posts",
+ json=payload
+ )
+
+ return response
+
+
+def preview_migration(html_path):
+ """Preview what would be migrated without posting."""
+ sections = parse_news_html(html_path)
+
+ print("\n" + "="*60)
+ print("MIGRATION PREVIEW - news.html to Tumblr (NPF Format)")
+ print("="*60)
+
+ total_posts = 0
+ for section in sections:
+ print(f"\n## {section['month_title']} (backdate: {section['backdate']})")
+ print("-" * 40)
+
+ for i, item in enumerate(section['items'], 1):
+ # Convert HTML to NPF for preview
+ html_content = "\n".join(item['html_parts'])
+ npf_blocks = html_to_npf_blocks(html_content)
+
+ text_blocks = sum(1 for b in npf_blocks if b['type'] == 'text')
+ image_blocks = sum(1 for b in npf_blocks if b['type'] == 'image')
+
+ print(f" {i}. {item['title'][:50]}")
+ print(f" Blocks: {len(npf_blocks)} ({text_blocks} text, {image_blocks} images)")
+ total_posts += 1
+
+ print("\n" + "="*60)
+ print(f"TOTAL: {total_posts} posts would be created")
+ print("="*60)
+
+ return sections
+
+
+def preview_single(html_path, section_id, item_index):
+ """Preview NPF blocks for a single item."""
+ sections = parse_news_html(html_path)
+
+ for section in sections:
+ if section['section_id'] == section_id:
+ if item_index < len(section['items']):
+ item = section['items'][item_index]
+ print(f"\n=== {item['title']} ===\n")
+
+ html_content = "\n".join(item['html_parts'])
+ print("--- HTML Input ---")
+ print(html_content[:1000])
+
+ print("\n--- NPF Output ---")
+ npf_blocks = html_to_npf_blocks(html_content)
+ print(json.dumps(npf_blocks, indent=2))
+ return
+
+ print(f"Section {section_id} or item {item_index} not found")
+
+
+def run_migration(html_path, state="draft", confirm=False):
+ """
+ Run the migration.
+
+ Args:
+ html_path: Path to news.html
+ state: 'draft' or 'published'
+ confirm: If False, just preview. If True, actually post.
+ """
+ sections = parse_news_html(html_path)
+ oauth = get_oauth_session()
+
+ if not confirm:
+ print("\n[DRY RUN] Add --confirm to actually create posts\n")
+
+ created = 0
+ failed = 0
+
+ for section in sections:
+ print(f"\n## {section['month_title']}")
+
+ for item in section['items']:
+ title = f"{section['month_title']}: {item['title']}"
+
+ # Convert HTML to NPF
+ html_content = "\n".join(item['html_parts'])
+ npf_blocks = html_to_npf_blocks(html_content)
+
+ # Default tags
+ tags = ["openworm", "c. elegans", "computational biology", "open science"]
+
+ if confirm:
+ response = create_npf_post(
+ oauth, title, npf_blocks,
+ section['backdate'],
+ state=state,
+ tags=tags
+ )
+
+ if response.status_code in [200, 201]:
+ result = response.json()
+ post_id = result.get('response', {}).get('id')
+ print(f" ✓ Created: {item['title'][:40]}... (ID: {post_id})")
+ created += 1
+ else:
+ print(f" ✗ FAILED: {item['title'][:40]}...")
+ print(f" Error: {response.status_code} - {response.text[:200]}")
+ failed += 1
+ else:
+ print(f" [would create] {title[:50]}... ({len(npf_blocks)} blocks)")
+
+ print(f"\n{'='*60}")
+ if confirm:
+ print(f"Created: {created} | Failed: {failed}")
+ else:
+ print("DRY RUN complete. Use --confirm to actually create posts.")
+ print(f"{'='*60}")
+
+
+if __name__ == "__main__":
+ import sys
+
+ html_path = Path(__file__).parent / "news.html"
+
+ if len(sys.argv) < 2:
+ print("""
+News to Tumblr Migration Tool (NPF Format)
+
+Usage:
+ python migrate_news_to_tumblr.py preview - See what would be migrated
+ python migrate_news_to_tumblr.py inspect SECTION IDX - Inspect a single item's NPF conversion
+ (e.g., inspect September2022 0)
+ python migrate_news_to_tumblr.py draft - Create as drafts (dry run)
+ python migrate_news_to_tumblr.py draft --confirm - Actually create drafts
+ python migrate_news_to_tumblr.py publish --confirm - Publish directly (careful!)
+ """)
+ sys.exit(0)
+
+ command = sys.argv[1].lower()
+ confirm = "--confirm" in sys.argv
+
+ if command == "preview":
+ preview_migration(html_path)
+ elif command == "inspect":
+ if len(sys.argv) >= 4:
+ section_id = sys.argv[2]
+ item_idx = int(sys.argv[3])
+ preview_single(html_path, section_id, item_idx)
+ else:
+ print("Usage: inspect SECTION_ID ITEM_INDEX")
+ print("Sections: June2025, Dec2024, May2024, June2023, September2022")
+ elif command == "draft":
+ run_migration(html_path, state="draft", confirm=confirm)
+ elif command == "publish":
+ if confirm:
+ print("\n⚠️ WARNING: This will PUBLISH posts directly!")
+ answer = input("Type 'yes' to continue: ")
+ if answer.lower() != 'yes':
+ print("Aborted.")
+ sys.exit(0)
+ run_migration(html_path, state="published", confirm=confirm)
+ else:
+ print(f"Unknown command: {command}")
+ sys.exit(1)
diff --git a/news.html b/news.html
index 5083689..43e36c8 100644
--- a/news.html
+++ b/news.html
@@ -5,7 +5,7 @@
OpenWorm News
-
+
@@ -16,11 +16,6 @@
-
-
-
@@ -55,277 +50,54 @@ OpenWorm News
Recent updates from the OpenWorm project
+
+ View full blog on Tumblr »
+
-
-
-
-
-
-
-
+
-
-
+
+
-
-
-
-
+
+
-
-
-
+
+
+
+
+
diff --git a/news.html.backup b/news.html.backup
new file mode 100644
index 0000000..3457b04
--- /dev/null
+++ b/news.html.backup
@@ -0,0 +1,327 @@
+
+
+
+
+
+ OpenWorm News
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+ OpenWorm News
+
+ Recent updates from the OpenWorm project
+
+
+
+
+
+
+
+
+
+
+
+
+
+ June 2025
+
+ 1) OpenWorm.ai - a C. elegans specific LLM
+
+ We are investigating the use of customized LLMs to constrain and validate computational models of C. elegans.
+ A prototype of this platform has been made available at openworm.ai. The core code for this can be found here.
+
+
+
+ 2) Updated C. elegans Connectome Toolbox
+
+ There have been significant updates to the C. elegans Connectome Toolbox (cect), with new connectomic datasets, and interactive views. It has also been more deeply integrated with the c302 package to allow that package to use any connectome from cect as a basis of computational models in NeuroML.
+
+
+
+
+
+
+
+ December 2024
+
+ 1) C. elegans Connectome Toolbox
+
+ A new package has been developed to help manage the multiple datasets related to C. elegans connectomics (e.g. chemical synapses, electrical connections, extrasynaptic, functional connectome, etc.).
+ Check out the C. elegans Connectome Toolbox at: https://openworm.org/ConnectomeToolbox.
+
+
+
+
+
+
+
+
+ May 2024
+
+ 1) OpenWorm is hiring!
+
+ A position for a Research Fellow or Senior Research Fellow at University College London is now open for applications. The successful applicant will contribute to multiple areas of the OpenWorm project, working closely with Padraig Gleeson. See here for more details.
+
+ 2) OpenWorm interviewed on Data Skeptic Podcast
+
+ Stephen Larson, Executive Director of the OpenWorm project, discusses the current state of the project as well as where our quest to develop a biologically realistic digital life form fits into the current landscape of AI. Listen to it here on Spotify.
+
+ 3) OpenWorm Docker simulation stack updated
+
+ A major update to the simulation stack of the OpenWorm project was released. This involved updating all core elements (c302, Sibernetic) in the Dockerfile and enabling automated tests for building the Docker container using Intel and AMD libraries. A big thanks to Austin Klein who completed this work as part of an OpenWorm Studentship.
+
+
+ 4) New cell and ion channel models in NeuroML
+
+ A key part of the OpenWorm project's work is to incorporate models of neurons which behave like their biological counterparts. The 2019 paper of Nicoletti and colleagues (Biophysical modeling of C. elegans neurons: Single ion currents and whole-cell dynamics of AWCon and RMD presents new models of 2 C. elegans neurons with the ionic currents which underlie their electrical activity. These have been converted to NeuroML format for use in OpenWorm. See here for full details.
+
+ 5) DevoWorm updates
+
+ 
+
+ DevoWorm celebrated its 10th anniversary in April. Bradly Alicea has prepared a 90-minute video on our progress over the past 10 years: from progress in OpenWorm to progress in computational developmental biology. If you would like to know more, please check out our weekly meetings on YouTube.
+
+ DevoWorm has sponsored two students through the Google Summer of Code program for 2024. Congrats to Pakhi Banchalia and Mehul Arora for being accepted to work on the Developmental Graph Neural Networks (D-GNNs) project. Pakhi will be working on incorporating Neural Developmental Programs (NDPs) into GNN models. Mehul will be working on hypergraph techniques for developmental lineage trees and embryogenesis. Himanshu Chougule, Google Summer of Code scholar for 2023, is a co-mentor for this project. Follow or fork the DevoGraph repository to contribute or keep up with our progress.
+
+
+
+
+
+
+
+ June 2023
+
+ 1) OpenWorm @ C. elegans 2023
+
+ The OpenWorm team were present at the main C. elegans scientific conference in Glasgow in June 2023.
+
+ We presented an update on the recent activities in the project (click on the poster below) and connected with a number of groups who are acquiring leading edge experimental data which can be used to help build and constrain our worm models.
+
+
+
+ 2) Hodgkin Huxley interactive tutorial
+
+ As part of a recent Google Summer of Code project by Rahul Sonkar, an interactive tutorial for the Hodgkin Huxley model of neuronal activity was developed.
+ This Jupyter notebook based tutorial can be used to investigate how ion channel dynamical properties underlie the generation of the action potential in neurons.
+
+
+
+ 3) Updated 2D worm body model visualisations
+
+ We have been investigating multiple options for incorporating 2D worm body models into our simulation pipeline to complement the full 3D Sibernetic-based worm body model.
+
+ Options investigated so far include the model of Olivares, Izquierdo & Beer, as well as
+ the Boyle, Berri and Cohen model, for which we have added a new Python based visualisation tool:
+
+
+
+
+ 4) DevoWorm updates
+
+ 
+
+ DevoWorm has sponsored two projects through the Google Summer of Code program for 2023: maintenance of the DevoLearn software package, and development of work on GNN (Graph neural Networks) and TDA (Topological Data Analysis) applications to biological development. These projects involve the work of GSoC scholars Sushmanth Reddy and Himanshu Chougule, respectively.
+
+ Members of the DevoWorm group have two recent publications: Embodied Cognitive Morphogenesis as a Route to Intelligent Systems, published at Royal Society Interface Focus, and The Psychophysical World of the Motile Diatom Bacillaria Paradoxa, published in Mathematical Biology of Diatoms.
+
+ In our weekly meetings, we have discussed topics such as early life and the origins of embryos, the biophysical and topological underpinnings of phenotypes, current organoid, embryoid, and assembloid research, and the intersection of microscopy and artificial life. There are collective discussions, as well as presentations on various topics by Bradly Alicea. Our archive of weekly meetings is available on the DevoWorm YouTube channel.
+
+
+
+
+
+September 2022
+
+1) OpenCollective as a new funding stream
+
+
+
+We have added OpenCollective as a new and easy way to donate to OpenWorm.
+ Regular contributions to the project (monthly/yearly) are possible through this funding stream.
+ See here for more details.
+
+One of the first uses of these funds will be to support OpenWorm Studentships (see below).
+
+
+2) OpenWorm Studentships - first successful project completed
+
+OpenWorm Studentships are a new way to incentivize contribution to the OpenWorm project,
+ offering small stipends and recognition to junior researchers who want to spend time bringing their research
+ into OpenWorm and making it more accessible for the wider community. See
+ here for more information.
+
+The first person to complete an OpenWorm Studentship project has been Tyson Wheelwright. He has made significant updates to
+ the Blender 2 NeuroML subproject.
+
+ +
+
+Figure showing a single cell (ADAL) highlighted in orange in the original
+3D Blender file (from the
+Virtual Worm Project) on the left, and corresponding images of the cell on its own in Blender (middle 2 panels) and in
+NeuroML 2 format (right 2 panels). The NeuroML version is being used in our
+biophysical model of the worm nervous system. See
+here for more examples.
+
+More Studentships will be offered to the community in late 2022.
+
+
+3) NeuroPAL
+
+The recent paper NeuroPAL: A Multicolor Atlas for Whole-Brain Neuronal Identification in C. elegans described a groundbreaking new technique to create a genetic strain of the worm where each neuron
+is labelled with a fluorescent marker of a specific color, allowing easy identification of neurons across experiments and animals.
+
+We have converted one of these datasets containing positions of cell bodies and the expressed NeuroPAL colors to NeuroML,
+ to allow it to be used in OpenWorm models and associated applications. Full details can be found
+ here.
+
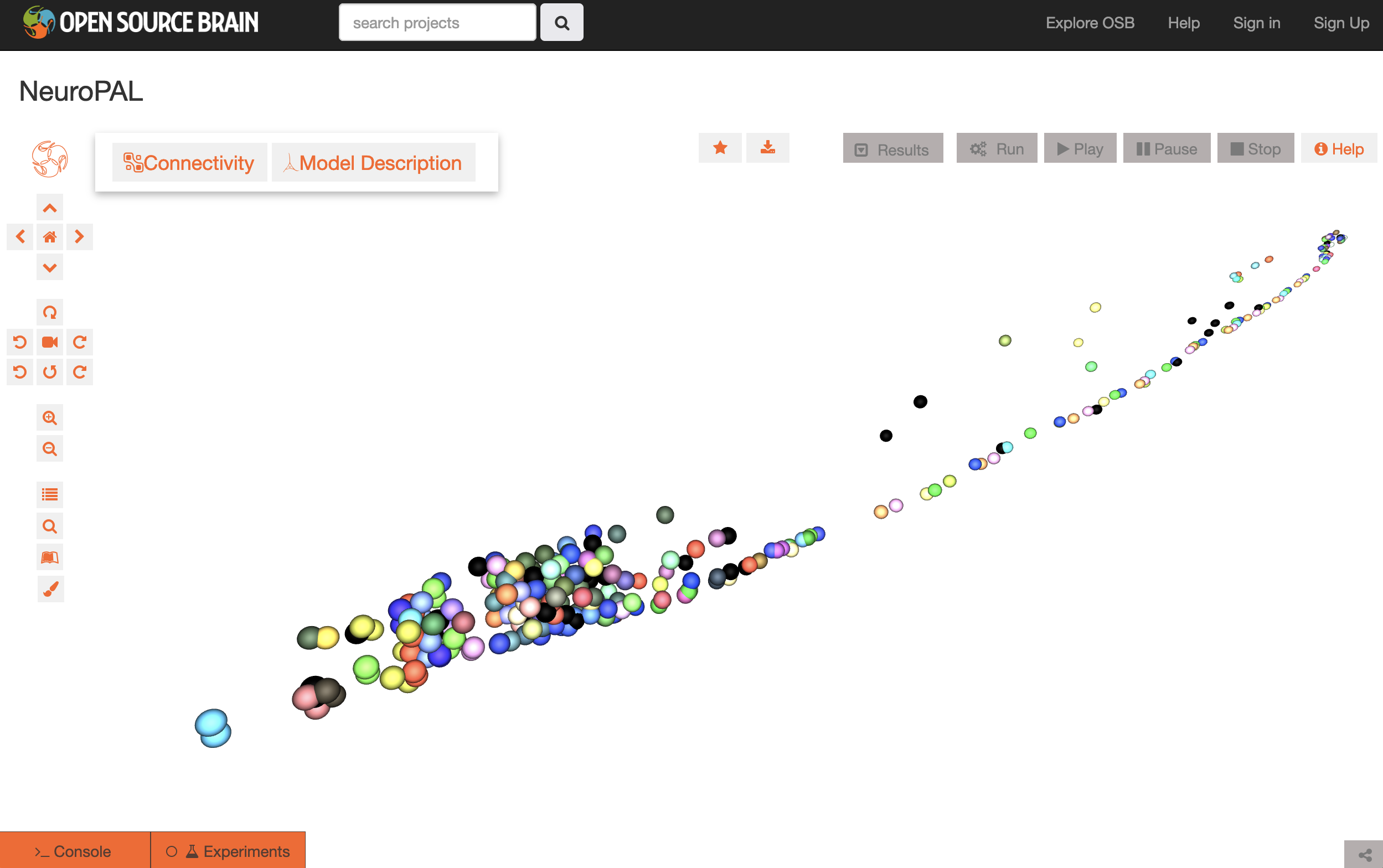
+
+
+The above image shows the canonical positions and colors of a NeuroPAL dataset which has been converted into NeuroML, allowing it to be visualised on Open Source Brain.
+
+
+4) Electrophysiological data from worm neurons
+
+Electrical recordings from individual neurons in C. elegans are crucial experimental data required for
+ creating biologically realistic computational models of the worm.
+
+Electrophysiological recordings of the activity of the
+ ASH neuron made by the
+ Wormsense Lab of Miriam Goodman were converted to open, standardized
+ Neurodata Without Borders format as part of a
+ Google Summer of Code project in 2021 by Steph Prince. Further details,
+ as well as all of the converted datasets, can be found
+ here.
+
+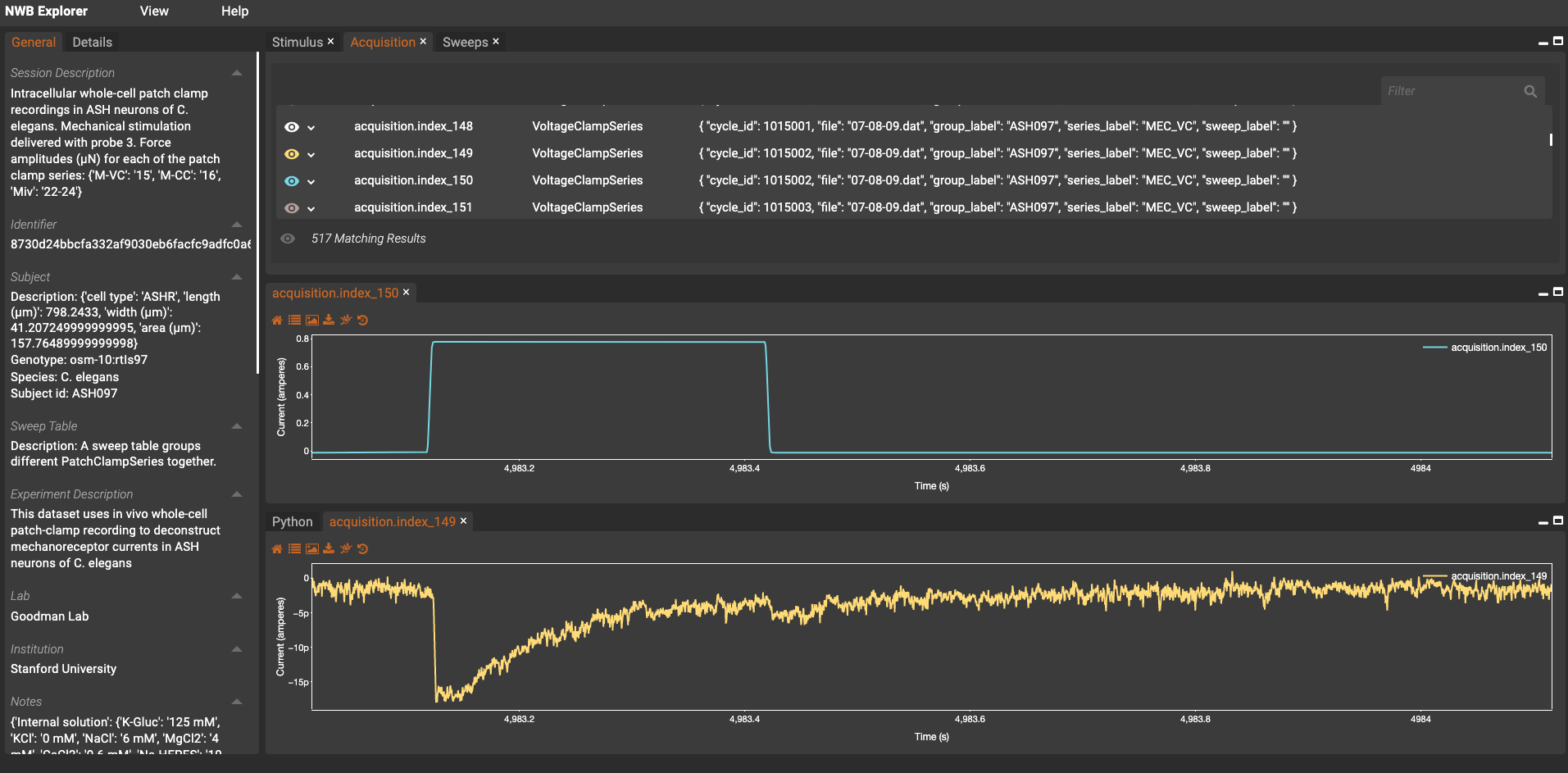
+A view of the
+NWB Explorer interface showing one of the converted electrophysiological datasets. Click
+here to explore other datasets and get direct
+links to NWB Explorer to browse them.
+
+
+5) DevoWorm updates
+
+
+
+Over the past year, DevoWorm has been involved in a number of initiatives.
+ DevoWorm hosted four Google Summer of Code students in 2022: Karan Lohaan and Harikrishna
+ Pillai worked on the Digital Microspheres project (a spherical platform for embryo data), while Jiahang Li and Wataru Kawakami worked
+ on developing a Graph Neural Network pipeline for the DevoLearn platform.
+
+DevoWorm also had a presence at NetSci 2022 (virtual conference hosted in Shanghai).
+ Bradly Alicea presented on the group's Embodied Hypernetworks work in the main conference, as well as serving to represent the group
+ at the Network Neuroscience satellite session as a presenter and participant.
+
+Our weekly meetings are a continuing success.
+ We cover a number of technical topics and a wide variety of topical reviews spanning biophysics, developmental biology, computational
+ analysis, and more. Join us on Mondays at 2pm UTC (1pm UTC in winter) to get involved.
+6) OpenWorm simulation stack updates
+
+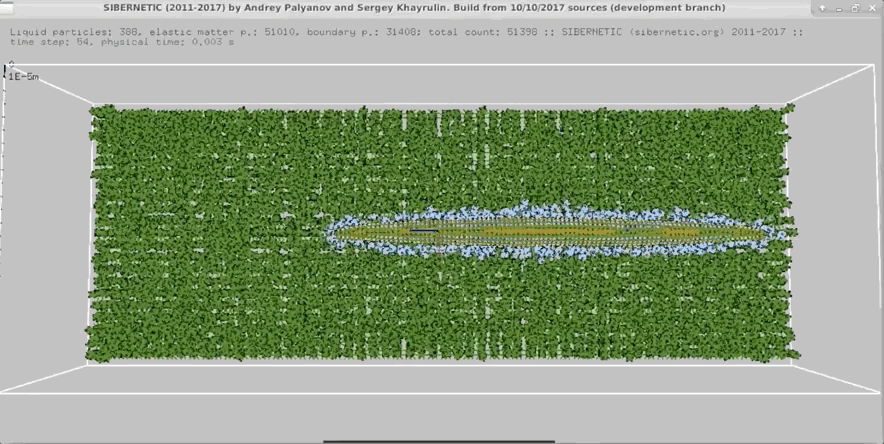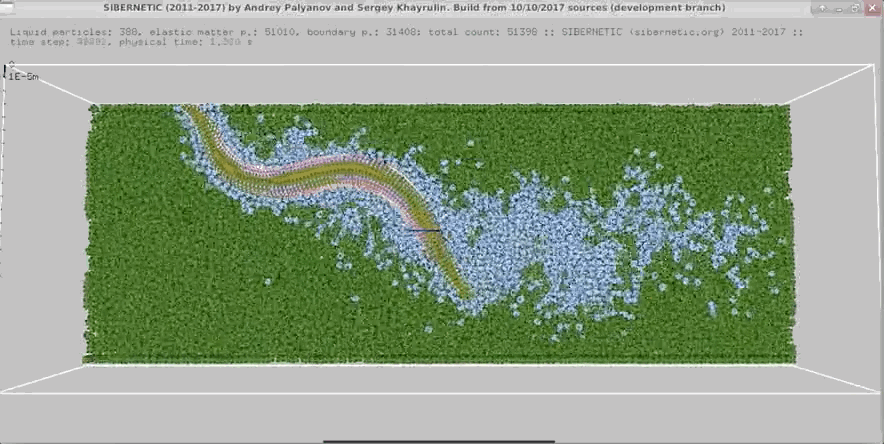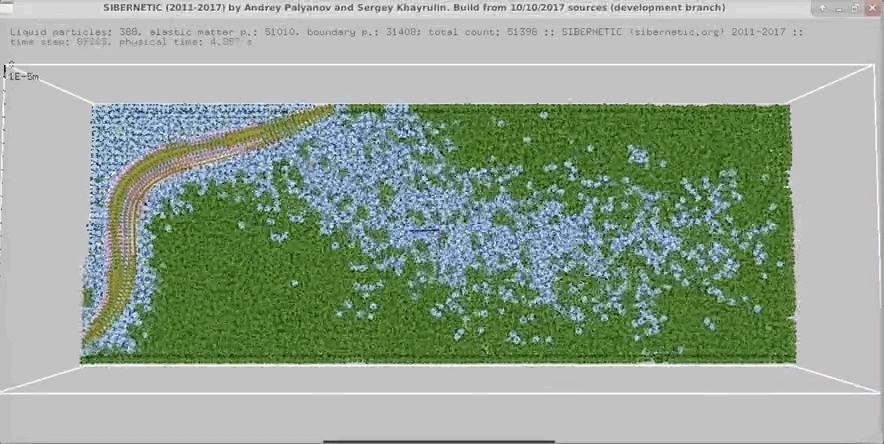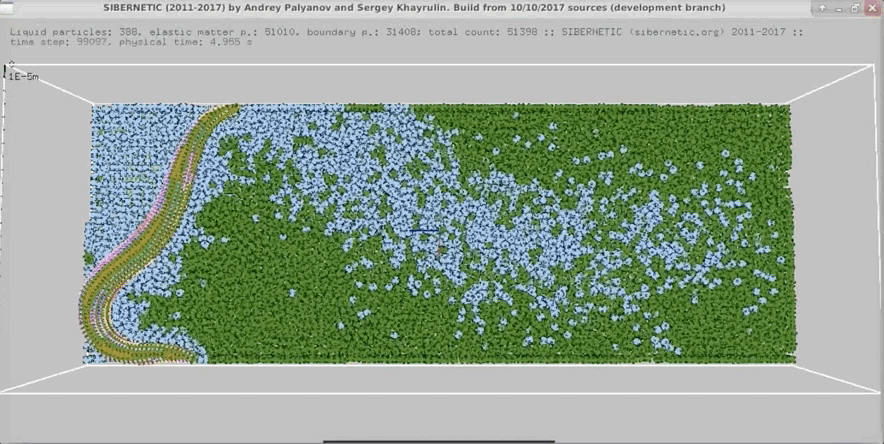
+Example movie generated when the Docker container containing the OpenWorm simulation stack runs for 5
+ seconds of simulated time.
+
+We have made a new release of the
+ Docker container that pulls together
+ the major components of our simulation stack and allows it to be run on your machine. It includes
+ Sibernetic,
+ c302 and
+ owmeta.
+ See details here.
+
+7) Other announcements
+
+
+
+A poster presenting the latest status of the OpenWorm project was presented at the
+ main global
+ conference in the C. elegans field in June 2021. Click on the image above to download the poster.
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
+
diff --git a/people.html b/people.html
index 2353994..bc3d347 100644
--- a/people.html
+++ b/people.html
@@ -16,11 +16,6 @@
-
-
-
@@ -939,7 +934,7 @@ Ryan Merkley
-
+
diff --git a/publications.html b/publications.html
index 68a05dc..932537d 100644
--- a/publications.html
+++ b/publications.html
@@ -15,11 +15,6 @@
-
-
-
@@ -479,7 +474,7 @@ Managing Complexity in Multi-Algorithm, Multi-Scale Biological Simulations:
-
+
diff --git a/repositories.html b/repositories.html
index 58b2244..afe9fd1 100644
--- a/repositories.html
+++ b/repositories.html
@@ -20,13 +20,8 @@
-
-
-
-
-
+
+
@@ -207,7 +202,7 @@ Repositories
-
+
@@ -215,7 +210,7 @@ Repositories
-
+
diff --git a/science.html b/science.html
index 34d0ca1..943d75d 100755
--- a/science.html
+++ b/science.html
@@ -15,11 +15,6 @@
-
-
-
@@ -106,9 +101,9 @@ Existing Scientific Collaborations
The entire C. elegans experimental community that assembles at the yearly
GSA C. elegans meeting
is incredibly welcoming and supportive. We've received data and help from C. elegans biologists including
- Dr. Sreekanth Chalasani at the Salk Institute,
+ Dr. Sreekanth Chalasani at the Salk Institute,
Dr. Michael Francis at UMass Medical School,
- Dr. William Schafer at University of Cambridge
+ Dr. William Schafer at University of Cambridge
and Dr. Andrew Leifer at Princeton University. We've been
inspired and received help on worm simulations from the lab of Dr. Netta Cohen at Leeds
University and her students. The original source for many of the 3D images of the worm's body
@@ -130,7 +125,7 @@ OpenWorm Documentation

OpenWorm News
++ Recent updates from the OpenWorm project +
+June 2025
+ +1) OpenWorm.ai - a C. elegans specific LLM
+ +We are investigating the use of customized LLMs to constrain and validate computational models of C. elegans. + A prototype of this platform has been made available at openworm.ai. The core code for this can be found here.
+ + + +2) Updated C. elegans Connectome Toolbox
+ +There have been significant updates to the C. elegans Connectome Toolbox (cect), with new connectomic datasets, and interactive views. It has also been more deeply integrated with the c302 package to allow that package to use any connectome from cect as a basis of computational models in NeuroML.
+ + + +December 2024
+ +1) C. elegans Connectome Toolbox
+ +A new package has been developed to help manage the multiple datasets related to C. elegans connectomics (e.g. chemical synapses, electrical connections, extrasynaptic, functional connectome, etc.). + Check out the C. elegans Connectome Toolbox at: https://openworm.org/ConnectomeToolbox.
+ + + + +May 2024
+ +1) OpenWorm is hiring!
+ +A position for a Research Fellow or Senior Research Fellow at University College London is now open for applications. The successful applicant will contribute to multiple areas of the OpenWorm project, working closely with Padraig Gleeson. See here for more details.
+ +2) OpenWorm interviewed on Data Skeptic Podcast
+ +Stephen Larson, Executive Director of the OpenWorm project, discusses the current state of the project as well as where our quest to develop a biologically realistic digital life form fits into the current landscape of AI. Listen to it here on Spotify.
+ +3) OpenWorm Docker simulation stack updated
+ +A major update to the simulation stack of the OpenWorm project was released. This involved updating all core elements (c302, Sibernetic) in the Dockerfile and enabling automated tests for building the Docker container using Intel and AMD libraries. A big thanks to Austin Klein who completed this work as part of an OpenWorm Studentship.
+ + +4) New cell and ion channel models in NeuroML
+ +A key part of the OpenWorm project's work is to incorporate models of neurons which behave like their biological counterparts. The 2019 paper of Nicoletti and colleagues (Biophysical modeling of C. elegans neurons: Single ion currents and whole-cell dynamics of AWCon and RMD presents new models of 2 C. elegans neurons with the ionic currents which underlie their electrical activity. These have been converted to NeuroML format for use in OpenWorm. See here for full details.
+ +5) DevoWorm updates
+ +
DevoWorm celebrated its 10th anniversary in April. Bradly Alicea has prepared a 90-minute video on our progress over the past 10 years: from progress in OpenWorm to progress in computational developmental biology. If you would like to know more, please check out our weekly meetings on YouTube.
+ +DevoWorm has sponsored two students through the Google Summer of Code program for 2024. Congrats to Pakhi Banchalia and Mehul Arora for being accepted to work on the Developmental Graph Neural Networks (D-GNNs) project. Pakhi will be working on incorporating Neural Developmental Programs (NDPs) into GNN models. Mehul will be working on hypergraph techniques for developmental lineage trees and embryogenesis. Himanshu Chougule, Google Summer of Code scholar for 2023, is a co-mentor for this project. Follow or fork the DevoGraph repository to contribute or keep up with our progress.
+ + + +June 2023
+ +1) OpenWorm @ C. elegans 2023
+ +The OpenWorm team were present at the main C. elegans scientific conference in Glasgow in June 2023.
+ +We presented an update on the recent activities in the project (click on the poster below) and connected with a number of groups who are acquiring leading edge experimental data which can be used to help build and constrain our worm models.
+ + + +2) Hodgkin Huxley interactive tutorial
+ +As part of a recent Google Summer of Code project by Rahul Sonkar, an interactive tutorial for the Hodgkin Huxley model of neuronal activity was developed. + This Jupyter notebook based tutorial can be used to investigate how ion channel dynamical properties underlie the generation of the action potential in neurons.
+ + + +3) Updated 2D worm body model visualisations
+ +We have been investigating multiple options for incorporating 2D worm body models into our simulation pipeline to complement the full 3D Sibernetic-based worm body model.
+ +Options investigated so far include the model of Olivares, Izquierdo & Beer, as well as + the Boyle, Berri and Cohen model, for which we have added a new Python based visualisation tool: + +
+ + +4) DevoWorm updates
+ +
DevoWorm has sponsored two projects through the Google Summer of Code program for 2023: maintenance of the DevoLearn software package, and development of work on GNN (Graph neural Networks) and TDA (Topological Data Analysis) applications to biological development. These projects involve the work of GSoC scholars Sushmanth Reddy and Himanshu Chougule, respectively.
+ +Members of the DevoWorm group have two recent publications: Embodied Cognitive Morphogenesis as a Route to Intelligent Systems, published at Royal Society Interface Focus, and The Psychophysical World of the Motile Diatom Bacillaria Paradoxa, published in Mathematical Biology of Diatoms.
+ +In our weekly meetings, we have discussed topics such as early life and the origins of embryos, the biophysical and topological underpinnings of phenotypes, current organoid, embryoid, and assembloid research, and the intersection of microscopy and artificial life. There are collective discussions, as well as presentations on various topics by Bradly Alicea. Our archive of weekly meetings is available on the DevoWorm YouTube channel.
+ +September 2022
+ +1) OpenCollective as a new funding stream
+ +
We have added OpenCollective as a new and easy way to donate to OpenWorm. + Regular contributions to the project (monthly/yearly) are possible through this funding stream. + See here for more details.
+ +One of the first uses of these funds will be to support OpenWorm Studentships (see below).
+ + +2) OpenWorm Studentships - first successful project completed
+ +OpenWorm Studentships are a new way to incentivize contribution to the OpenWorm project, + offering small stipends and recognition to junior researchers who want to spend time bringing their research + into OpenWorm and making it more accessible for the wider community. See + here for more information.
+ +The first person to complete an OpenWorm Studentship project has been Tyson Wheelwright. He has made significant updates to + the Blender 2 NeuroML subproject.
+ + +
+
Figure showing a single cell (ADAL) highlighted in orange in the original +3D Blender file (from the +Virtual Worm Project) on the left, and corresponding images of the cell on its own in Blender (middle 2 panels) and in +NeuroML 2 format (right 2 panels). The NeuroML version is being used in our +biophysical model of the worm nervous system. See +here for more examples.
+ +More Studentships will be offered to the community in late 2022.
+ + +3) NeuroPAL
+ +The recent paper NeuroPAL: A Multicolor Atlas for Whole-Brain Neuronal Identification in C. elegans described a groundbreaking new technique to create a genetic strain of the worm where each neuron +is labelled with a fluorescent marker of a specific color, allowing easy identification of neurons across experiments and animals.
+ +We have converted one of these datasets containing positions of cell bodies and the expressed NeuroPAL colors to NeuroML, + to allow it to be used in OpenWorm models and associated applications. Full details can be found + here.
+ +
The above image shows the canonical positions and colors of a NeuroPAL dataset which has been converted into NeuroML, allowing it to be visualised on Open Source Brain.
+ + +4) Electrophysiological data from worm neurons
+ +Electrical recordings from individual neurons in C. elegans are crucial experimental data required for + creating biologically realistic computational models of the worm.
+ +Electrophysiological recordings of the activity of the + ASH neuron made by the + Wormsense Lab of Miriam Goodman were converted to open, standardized + Neurodata Without Borders format as part of a + Google Summer of Code project in 2021 by Steph Prince. Further details, + as well as all of the converted datasets, can be found + here.
+ +
A view of the +NWB Explorer interface showing one of the converted electrophysiological datasets. Click +here to explore other datasets and get direct +links to NWB Explorer to browse them.
+ + +5) DevoWorm updates
+ +
Over the past year, DevoWorm has been involved in a number of initiatives. + DevoWorm hosted four Google Summer of Code students in 2022: Karan Lohaan and Harikrishna + Pillai worked on the Digital Microspheres project (a spherical platform for embryo data), while Jiahang Li and Wataru Kawakami worked + on developing a Graph Neural Network pipeline for the DevoLearn platform.
+ +DevoWorm also had a presence at NetSci 2022 (virtual conference hosted in Shanghai). + Bradly Alicea presented on the group's Embodied Hypernetworks work in the main conference, as well as serving to represent the group + at the Network Neuroscience satellite session as a presenter and participant.
+ +Our weekly meetings are a continuing success. + We cover a number of technical topics and a wide variety of topical reviews spanning biophysics, developmental biology, computational + analysis, and more. Join us on Mondays at 2pm UTC (1pm UTC in winter) to get involved.
+6) OpenWorm simulation stack updates
+ +
Example movie generated when the Docker container containing the OpenWorm simulation stack runs for 5 + seconds of simulated time.
+ +We have made a new release of the + Docker container that pulls together + the major components of our simulation stack and allows it to be run on your machine. It includes + Sibernetic, + c302 and + owmeta. + See details here.
+ +7) Other announcements
+ + + +A poster presenting the latest status of the OpenWorm project was presented at the + main global + conference in the C. elegans field in June 2021. Click on the image above to download the poster.
+ +Ryan Merkley
-
+
diff --git a/publications.html b/publications.html
index 68a05dc..932537d 100644
--- a/publications.html
+++ b/publications.html
@@ -15,11 +15,6 @@
-
-
-
@@ -479,7 +474,7 @@ Managing Complexity in Multi-Algorithm, Multi-Scale Biological Simulations:
-
+
diff --git a/repositories.html b/repositories.html
index 58b2244..afe9fd1 100644
--- a/repositories.html
+++ b/repositories.html
@@ -20,13 +20,8 @@
-
-
-
-
-
+
+
@@ -207,7 +202,7 @@ Repositories
Repositories
- All of the documentation for the project in a single site. + All of the documentation for the project in a single site.
OpenWorm Browser
@@ -152,7 +147,7 @@
NeuroML C. elegans Connectome

- The team has produced a model of the connectome of the C. elegans in NeuroML, + The team has produced a model of the connectome of the C. elegans in NeuroML, a standard for describing computational models of neurons and networks. The model has been generated by post-processing the morphology of the nervous system available in the Blender file produced by the Virtual Worm project with @@ -172,7 +167,7 @@
Sibernetic

- OpenWorm has been developing Sibernetic, a C++ implementation of the Predictive-corrective + OpenWorm has been developing Sibernetic, a C++ implementation of the Predictive-corrective Incompressible Smooth Particle Hydrodynamics (PCI-SPH) algorithm based on the work of Solenthaler. The algorithm has been extended to model contractile matter and membranes for simulating the muscle tissue of the C. elegans. The implementation uses OpenCL to distribute the computation @@ -186,28 +181,28 @@
Geppetto

- OpenWorm has been developing Geppetto, a web-based multi-algorithm, multi-scale simulation
+ OpenWorm has been developing Geppetto, a web-based multi-algorithm, multi-scale simulation
platform engineered to support the simulation of complex biological systems and their
surrounding environment. The platform aims at integrating multiscale models whose simulation
depends on a variety of algorithms which need to interoperate. Great focus has been put on
the engineering of the platform, aiming at having a robust and generic solution that can be
applied to different class of models. OpenWorm aims to make progresses of the OpenWorm
simulation available to scientist and general public through Geppetto. Geppetto at present
- supports a simulator for LEMS, a domain agnostic language specification aimed at describing
+ supports a simulator for LEMS, a domain agnostic language specification aimed at describing
dynamic models, and Sibernetic, the fluid dynamics simulator also developed as part of the
project. The visualisation engine of Geppetto has been reused in Open Source Brain Explorer
and allows a variety of neuronal models to be seamlessly visualised in 3D through the web
browser.
- Geppetto documentation
- Geppetto sources
- Geppetto website
+ Geppetto documentation
+ Geppetto sources
+ Geppetto website
Other activities
- OpenWorm is contributing the development of computational neuronal models to Open Source Brain. - The group page with all the projects can be found here. + OpenWorm is contributing the development of computational neuronal models to Open Source Brain. + The group page with all the projects can be found here.






